Version 1.0.4
The graphics functionality lost when using the Big Sur or higher operating systems has been restored.
Version 1.0.3
Minor update with a new support site and a privacy statement
Version 1.02
Minor update ensuring zero inhibition concentrations are removed before log values are obtained.
Version 1.01
The App has been updated to a 64 bit application.
Description
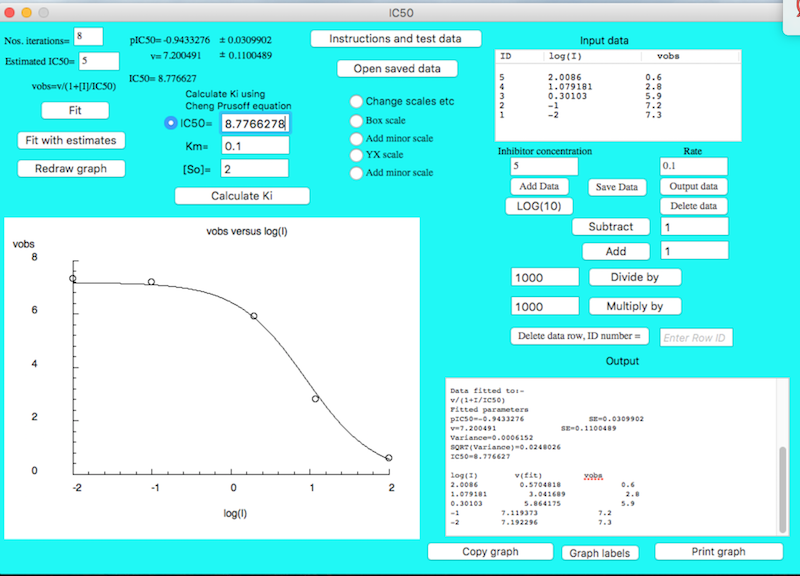
This program fits to vobs = v/(1+[I]/IC50) in the log form and so it gives pIC50 with standard errors. pIC50 is in the same format as a pKa i.e. it is -log(IC50). The Standard errors of pIC50 are log values and cannot be used to give a single standard error for the IC50 value.The best way to obtain errors for IC50 is to determine the IC50 values 3 or more times and determine the mean IC50 value and its standard deviation.
If competitive inhibition is occurring and if the substrate concentration used is very much smaller than its Km([So] <<Km) then the IC50 value will equal the Ki for the competitive enzyme inhibitor. If this approximation does not hold then you can use the Cheng-Prusoff equation to estimate the Ki value from the IC50 value.
Results can printed, saved as a text document or copied and pasted into other applications such as Word.
Specific instructions:-
FIRST INPUT DATA OR OPEN SAVED DATA
Buttons will be enabled as you need them.
Test data
I(mM)-------vobs
0.01————7.3
0.10————7.2
2.00————5.9
12.0————2.8
102————-0.6
Enter data into the boxes "log(I)" and "vobs"
Press button "Add Data"
Enter data into the boxes "log(I)" and "vobs"
Select button "Add Data"
Repeat until all data is entered.
A minimum of 3 different data pairs are required but preferably more points should be added.
Then you can save the data as a text file by selecting the "Save Data" button.
You can then reload the data at any time by selecting "Open Saved Data".
If you have data in a simple two column excel worksheet you can save it as a csv file. If you go to the file menu you will be able to open the csv file
You can delete all the data by selecting the "Delete data" button.
You can then reload the saved data at any time by selecting the "Open saved data" button.
To delete an individual data row you must first enter the ID number of the row you wish to delete in the box opposite the "Delete data row" button. When you click on the "Delete data row" button the row will be deleted and a new set of ID numbers will be generated.
Selecting the "output data" button outputs data to the output window.
TO ANALYSE DATA
Press LOG(10) to convert I values to log(I) values
Select the required fit. In most cases the option "Fit" will be suitable. Occasionally the initial linear fit will not give suitable initial estimates. In this case you should enter your own estimates and select the "Fit with estimates" option which will be available after you have used the "fit" option.
Answers for test data
pIC50=0.9433276 SE=0.0309902
k=7.200491 SE=0.1100489
IC50= 8.776627
After selecting the required fit the analyzed data, fitted data and the fitted parameters will be outputted to the Output window. This output can be selected, printed or copied and pasted into other applications such as word. These options will be found under the File and Edit menus.
Then select the type of scale you require.
Select "Change scales etc" and carefully enter the changes you require.
After changing the graph scales, press redraw to implement the changes.
To give the graph a title and label the axes, select "Graph labels" and make the appropriate changes.
After changing the graph scales or labels, press "redraw" to implement the changes.
Select "Print graph" to output the graph to your printer.
Select "Copy graph" to copy the graph to the clipboard. You may then paste it into other applications such as Word.
Calculating Ki values
If you are following the inhibition of the enzyme catalysed breakdown of a substrate by a competitive inhibitor then if the substrate concentration is very much smaller than the Km ([So]<<KM) of the substrate then the IC50 value will equal the Ki value.
If however the substrate concentration is not much lower than the Km then the IC50 value does not equal Ki. However, if you know the substrate concentration and its Km value then you can calculate Ki using the Cheng Prusoff equation. Simply enter the IC50, Km and substrate concentration and then press "Calculate Ki" and the Ki value will be printed in the Output window. This assumes that the enzyme concentration is negligible compared to the substrate and inhibitor concentrations.
For a more detailed discussion of the conversion of IC50 values to Ki values see
Cheng & Prusoff 1973 Biochem Pharmacol. 22(23):3099-108
Car et al., 2009 Nucleic Acids Res. 37(Web Server issue): W441–W445.
The analyzed data, fitted data and estimates of parameters will be outputted to the Output window. This output can be selected, printed or copied and pasted into other applications such as word. These options will be found under the File and Edit menus.
Select "Change scales. After changing the graph scales, press redraw to implement the changes.
You do not need to do anything for the test data. Simply close the "Change scales etc" window and then press "Redraw".
To give the graph a title and label the axes select "Graph labels" and make the appropriate changes.
AFTER CHANGING THE GRAPH SCALES, AXES OR LABELS PRESS "REDRAW" TO IMPLEMENT THE CHANGES.
Select "Print graph" to output the graph to your printer
Select "Copy graph" to copy the graph to the clipboard. You may then paste it into other applications such as Word.
If you have any questions please contact
